September 24, 2020
Mystery of giant proton pump solved
Mechanism how complex I of respiratory chain pumps protons finally understood – Study published in Science
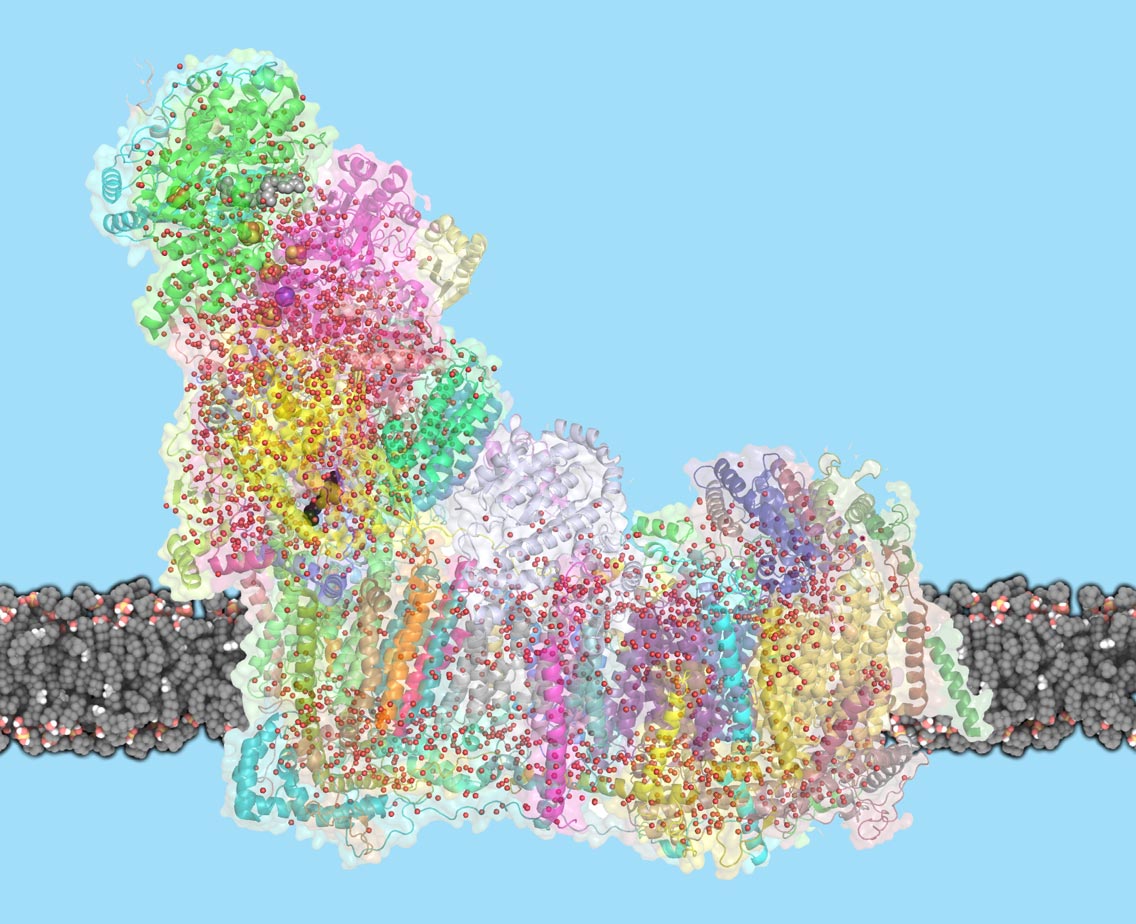
Mitochondria are the powerhouses of our cells, generating energy that supports life. A giant molecular proton pump, called complex I, is crucial: It sets in motion a chain of reactions, creating a proton gradient that powers the generation of ATP, the cell’s fuel. Despite complex I’s central role, the mechanism by which it transports protons across the membrane has so far been unknown. Now, Leonid Sazanov and his group at the Institute of Science and Technology Austria (IST Austria) have solved the mystery of how complex I works: Conformational changes in the protein combined with electrostatic waves move protons into the mitochondrial matrix. This is the result of a study published today in Science.
Complex I is the first enzyme in the respiratory chain, a series of protein complexes in the inner mitochondrial membrane. The respiratory chain is responsible for most of the cell’s energy production. In this chain, three membrane proteins set up a gradient of protons, moving them from the cell’s cytoplasm into the mitochondrial inner space, called the matrix. The energy for this process comes mostly from the electron transfer between NADH molecules, derived from the food we eat, and oxygen that we breathe. ATP synthase, the last protein in the chain, then uses this proton gradient to generate ATP. Complex I is remarkable not only because of its central role in life, but also for its sheer size: with a molecular weight of 1 Megadalton, the eukaryotic complex I is one of the biggest membrane proteins. Its size also makes complex I hard to study. In 2016, Sazanov and his group were the first to solve the structure of mammalian complex I, following on their 2013 structure of a simpler bacterial enzyme. But the mechanism by which complex I moves protons across the membrane has remained controversial. “One idea was that a part of complex I works like a piston, to open and close channels through which protons travel”, explains Sazanov. “Another idea was that residues at the center of complex I act as a driver. It turns out that an even more unusual mechanism is at work.”
Water wire helps protons to hop across the membrane
Previously, Sazanov and his group have shown that L-shaped complex I consists of hydrophilic and membrane arms. In the hydrophilic arm, electrons tunnel from NADH to quinone, the hydrophobic electron carrier. The membrane arm, where proton translocation happens, has three similar subunits with structures related to antiporters, and one subunit containing a quinone binding cavity. In this cavity, complex I transfers two electrons per catalytic cycle to quinone, which delivers the electrons further to complexes III and IV. But mystery surrounded how the interaction between electrons and quinone can move four protons per cycle across the membrane, since the antiporter-like subunits are far away from quinone cavity. To solve this puzzle, Sazanov and his team performed cryo-EM on sheep complex I. In a tour-de-force effort, PhD student Domen Kampjut solved 23 different structures of complex I, obtained under different conditions. By adding NADH and quinone, the researchers could capture images of complex I at work, changing conformation between the two main states. Due to high-resolution achieved, they could resolve the water molecules inside the protein, which are essential to allow proton transfer. They found that many water molecules in the central axis of the membrane arm provide a way for protons to hop between polar residues and waters, forming pathways along and across the membrane.
But only in one subunit, furthest away from quinone, do protons hop across the membrane. The other two subunits rather provide a coupling between the farthest subunit and quinone. When the binding cavity “waits” for quinone, a helix blocks the water wire in the central axis. When quinone binds in the binding cavity, the protein conformation around this area changes dramatically and this helix rotates. Now, the water wire connects all membrane subunits of complex I and two protons are delivered to quinone, to complete its reduction. This key part of the mechanism creates a charge near the first antiporter and starts an electrostatic wave of interactions between charged residues, which propagates along the antiporters, resulting in the translocation of four protons in total. “We show that a new and unexpected mechanism is at work in complex I. A mixture of both conformational changes and an electrostatic wave pumps protons across the membrane”, explains Sazanov. “This mechanism is highly unusual, as it involves the rotation of an entire helix inside the protein. It seems a bit excessive, but probably helps the mechanism to be robust.”
The new research complements studies from Sazanov group published in the last two months, on the mechanism of proton pumping in bacterial complex I (Nature Communications) and on the structure of MRP antiporters, from which complex I has evolved (eLife).



