September 14, 2022
Proton Dominos Kick Off Life
Universal mechanism of molecular machine complex I observed at ISTA
In a new Nature paper published today, ISTA professor Leonid Sazanov and his team propose a universal mechanism of cellular respiration, common in most species, thus culminating scientific work of around 20 years. These crucial findings in the field of bioenergetics are based on direct observation by cryogenic electron microscopy (cryo-EM) at the Institute of Science and Technology Austria (ISTA).
The extra-large molecular machine called complex I essentially kick-starts energetics within cells. Understanding how this elaborate proton pump functions is crucial to biology. It has been studied closely over the years, owing to its role as the entry point of the electron transport chain in cellular respiration. In a new paper released online today, Sazanov and his team delineate the mechanism of interaction between complex I’s different structural units. To harness energy for the cell, this is where electron transfer begins and protons are pumped across the membrane as a consequence, to energize the cell.
Leonid Sazanov, a prominent scientist in this field and one of the authors of the study heads the group “Structural Biology of Membrane Protein Complexes” that conducted this work at ISTA. Summarizing his recent findings, he says: “We have now provided a detailed explanation of each step in the mechanism. Every proton movement is accounted for. And it explains all the unusual features of complex I.”
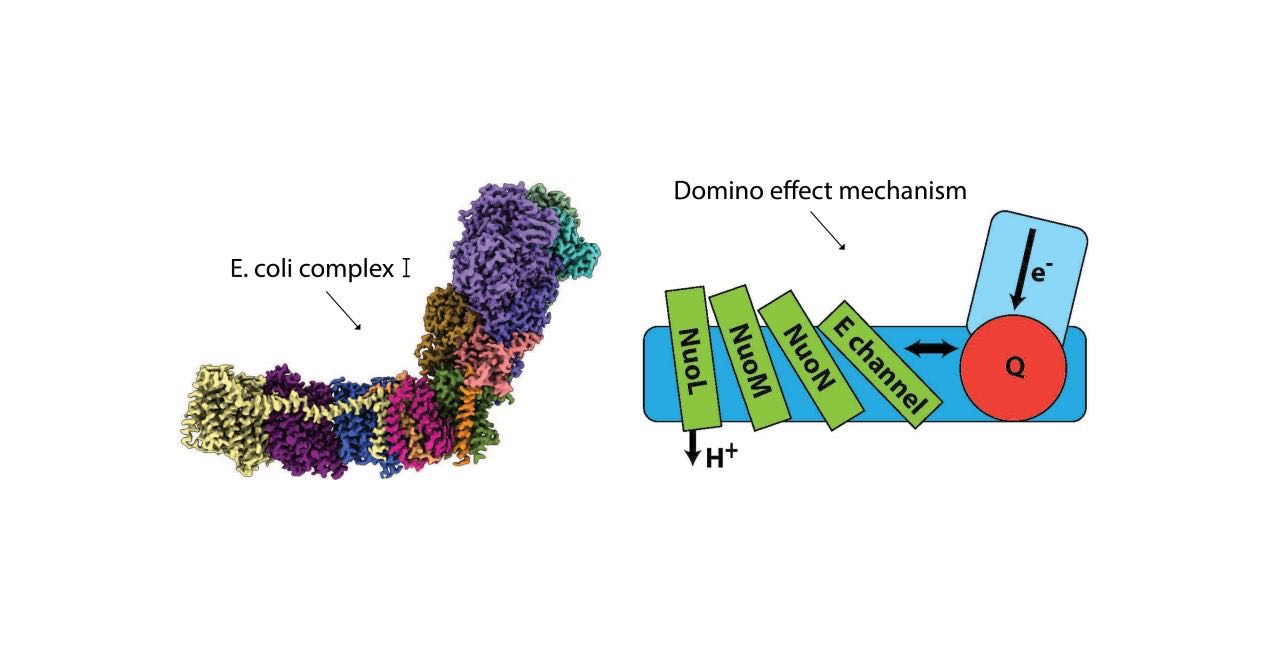
In the paper, Sazanov and his team propose a detailed yet robust mechanism comprising a “domino effect” of proton transfers and electrostatic interactions. A forward wave (dominoes stacking) primes the pump and the reverse wave (dominoes falling) results in the ejection of all pumped protons from a distal subunit of complex I. Previously, it was suggested that multiple subunits expel protons. The new work, however, observed that most of protons move along, rather than across the membrane, from one subunit into the next and ultimately leaving from the distal end of complex I membrane arm.
A Universal Mechanism
Importantly, the group designed their studies in such a way that the result would verify previous findings to reveal universal mechanistic principles in most living beings – from bacteria to mammals and also in molecular machines similar to complex I. In 2020, their work describing for the first time, the atomic structure and mechanisms of complex I in mammalian (ovine, or sheep) mitochondria was published. Mammalian complex I is a mega-molecule weighing around 1 megadalton (MDa) and in turn arranged as supercomplexes. The current work used E. coli, a bacterial model, where complex I weighs around 0.5 MDa and contains only conserved subunits.
From the structure of complex I it is known that the mega-structure can change its form: it either opens up its arms or tightly closes them. In the mammalian model, the Sazanov group saw both open and closed conformations and suggested that complex I cycles through them when it is working. That raised many questions, as different conformations were not observed in other species. To clarify all doubts, Sazanov says, the group decided to use E. coli next because of its large evolutionary distance to mammals. This time too, they report both open and closed states in E. coli, similar to mammalian states but with a key difference. The closed state was observed only under turnover (i.e. during catalysis), which shows that it really is a catalytic intermediate. “That has proven that open and closed states are both intermediate states in complex I’s activity,” Sazanov explains.
Watch the corresponding video on Youtube
“There are core protein subunits that are conserved from E. coli to mammals. Previously we described our preliminary mechanism in the mammalian system, which is slightly complicated due to all the extra subunits. But we have seen the same in E. coli, which means that our mechanism, now more detailed, is universal in all species with complex I,” Sazanov says.
This work not only confirms the pathbreaking findings in the 2020 paper but goes on to propose a universal “domino effect” mechanism of proton translocation that ultimately fires up our cells for growth, functions, and reproduction. “This is a universal coupling mechanism of complex I and related enzymes,” the authors contend in the paper that will appear in print in Nature on 22nd September.
New Benchmark for Cryo-EM
The state-of-the-art cryogenic electron microscope housed at ISTA makes these findings possible. With its help, the Sazanov group can routinely make observations approaching 2 Armstrong resolution. This is enough to see water molecules and thus identify the proton transfer pathways. Today, the cryo-EM technology is mature and fast enough to get enough data so as to develop mechanistic models of molecular machines. Each structure takes only about a month, which would be much longer with techniques such as X-ray crystallography.
Leonid Sazanov has been interested in large molecular machines since the beginning of his scientific career. When he began his work in the late 90s, the structure of L-shaped complex I was largely unknown. He subsequently worked for many years with X-ray crystallography to solve first the structure of the hydrophilic arm and then the membrane arm, whose exact functioning he has now uncovered. He confesses that this recent work could be approached with X-ray crystallography but would take perhaps 20-30 more years.
The first author of the study and Ph.D. student at the Sazanov lab, Vladyslav Kravchuk carried out most of the observations with the cryo-EM – a task that took about 3 years. More than 20 different datasets were collected that were necessary to observe complex I in action in different states, along with mutagenesis experiments that the group undertook. Each dataset consists of close to 5,000 micrographs taken usually over 24 hours. Micrographs from each dataset then provide about a million single-particle images, which are selected and analyzed computationally using ISTA high-performance computer cluster. “One of the main challenges of the project was careful analysis of cryo-EM data, as complex I is quite flexible and adopts different conformational states. Only with to the extensive, careful and sometimes non-conventional analysis we managed to fully resolve the closed state mentioned above. This clarified many questions and debates in the field and will further serve as a benchmark for cryo-EM data analysis for complex I and related molecules,” says Vladyslav Kravchuk.
Publication:
https://www.nature.com/articles/s41586-022-05199-7
Kravchuk, V. et al. 2022. A universal coupling mechanism of respiratory complex I. Nature.
DOI: 10.1038/s41586-022-05199-7
Funding information:
This research was supported by the Scientific Service Units (SSU) of ISTA through resources provided by the Electron Microscopy Facility (EMF), the Life Science Facility (LSF) and the IST high-performance computing cluster. First author was a recipient of a DOC Fellowship of the Austrian Academy of Sciences at the Institute of Science and Technology, Austria. Two of the authors are funded by the ERC Advanced Grant to Professor Leonid Sazanov.



